Abstract
During fetal development, hematopoietic stem cells (HSCs) undergo a remarkable expansion through a combination of rapid proliferation and high rates of self-renewal. In contrast, adult HSCs are characterized by long-term quiescence. Understanding of the molecular mechanisms underlying these ontogeny-dependent differences in cell cycle and self-renewal is hampered by marked heterogeneity within the HSC compartment, making it difficult to distinguish overlapping signatures in bulk transcriptional data. Advances in single cell genomics provide a new opportunity to tease apart different sources of gene expression heterogeneity, including those relating to cell cycle and self-renewal capability.
To address these questions, and improve resolution for cell-cycle annotation of individual HSCs, we developed an integrated single cell (sc)RNA-seq and live cell-cycle staining technique using Hoechst 33342 (DNA) and Pyronin-y (RNA) based FACS index sorting, followed by smart-seq2 based scRNA-seq. We validated our approach on 4 hematopoietic cell lines from mouse and human, using these data as a training set to apply a novel integrated pseudotime package that orders single cells by stage of cell cycle rather than developmental trajectory. By this approach we detected non-canonical cell cycle genes not apparent through bulk sorting of distinct cell cycle phases, and not previously annotated in published cell cycle gene sets (sc-pseudotime genes = 665, FDR < 0.05; non-annotated cell cycle = 487; non-bulk detected = 570).
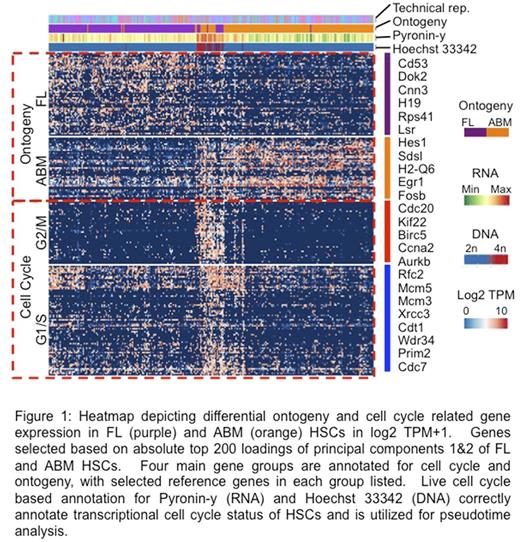
We then applied our technique to analyze primary mouse HSCs from 3 developmental time points (e15.5 fetal liver (FL), 2 week old bone marrow, and 6 week old adult bone marrow (ABM), n >1,500 single cells). Our cell cycle based integrated pseudotime analysis revealed distinct cell cycle signatures for FL, ABM, and common cell cycle related transcripts across distinct developmental time points that have not previously been described, including 555 unique cell cycle genes in FL; 401 unique cell cycle genes in ABM, and 93 novel cell cycle genes in common to both developmental time points e.g. Pclaf, Zfp367 and including long non-coding RNAs e.g. Lockd (FDR < 0.001).
Our dataset uniquely allowed us to explore ontogeny related molecular signatures without the overriding effect of confounding cell cycle associated gene expression by directly comparing non-mitotic cells from FL and ABM groups. We identified 404 differentially expressed transcripts (FDR <0.001, >2FC), including genes of unknown HSC function (e.g., FL: Lgals1, Gmfg; ABM: Zfp36l1, Rgs1). Single cell qPCR confirmed aberrant expression of 26/29 (89.7%) selected ontogeny candidate genes. Furthermore, hallmark gene set enrichment analysis revealed upregulation of oxidative phosphorylation, MYC targets, and E2F targets in FL; and TNFA signaling, Hypoxia, and TGF-beta signaling, among others, in ABM (FDR <0.01, NES > 1.5).
We then functionally reversed ABM quiescence through in vivo 5-FU treatment, and performed our single cell RNA-seq analysis on HSCs both 2 and 6 days post injection. This allowed us to identify genes and pathways associated with selective resistance of HSCs to chemotherapy, including upregulation of the hallmark gene sets for unfolded protein response, fatty acid metabolism, and MTOR signaling (FDR < 0.01, NES > 1.5 for each set).
To functionally validate novel ontogeny related genes we utilized genetic mouse models for two unexplored ABM related genes, Zfp36L1, an RNA-binding zinc finger protein, and Rgs1 a regulator of G-protein coupled signaling. We transplanted Cre-ERT2 conditionally floxed Zfp36L1 bone marrow with CD45.1 competitor control and induced deletion by tamoxifen four weeks post transplant. Compared to the initial four-week post transplant time point we observed a significant reduction in chimerism from Zfp36L1 deleted bone marrow compared to Cre-ERT2 control (p < .05). Competitive transplantation of Rgs1 -/- and WT bone marrow at a 1:1 ratio with CD45.1 competitors resulted in significantly reduced myeloid chimerism at 16 weeks post transplant. Secondary transplant and single cell cycle molecular analysis of these mice are ongoing together with functional validation of a number of other candidate genes.
Our results demonstrate the utility of single cell analysis to discover novel HSC regulators providing a unique dataset for further studies investigating regulators of HSC function.
Mead: BMS: Honoraria; Pfizer: Honoraria; Novartis: Honoraria, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.